lsd test package in r|fisher's least significant difference lsd : service This tutorial shows how to do the post hoc test using Fisher's LSD (Least Significant Different) in R. It includes data example and R code. Reinildo (born 21 January 1994) is a Mozambican footballer who plays as a left back for Spanish club Atlético Madrid. In the game FC 24 his overall rating is 81. Players
{plog:ftitle_list}
UX para escrita à mão digital natural. A escrita à mão entra no mundo digital com uma IA .
LSD.test function - RDocumentation. agricolae (version 1.3-7) LSD.test: Multiple comparisons, "Least significant difference" and Adjust P-values. Description. Multiple comparisons of . One commonly used post-hoc test is Fisher’s least significant difference (LSD) test. You can use the LSD.test() function from the agricolae package to perform this test in R. The following example shows how to use . library(agricolae) LSD_Test<- LSD.test(anova3way,"sowing_date") LSD_Test. For example, I'd like to check the mean difference under 3 way interaction, and also interaction between . Fisher’s Least Significant Difference (LSD) test is a powerful tool for comparing the means of more than two groups after conducting ANOVA. In this article, we have gone through the theoretical background, the .
This tutorial shows how to do the post hoc test using Fisher's LSD (Least Significant Different) in R. It includes data example and R code.For the mean comparison among variables, Least Significant difference (LSD) test is the most common method. Today I’ll introduce LSD test using R Studio. Here is one data.
Description. It performs the multivariate Left Spherically Distributed linear scores test of L\"auter et al. (The Annals of Statistics, 1998) (see also details below). Usage. lsd.test(resp, alternative .Use the lsd.test() function from the agricolae package to perform the LSD test. The syntax for the function is lsd.test(response ~ group, data = YourData), where "response" represents the variable you want to analyze, "group" means the .
the controversy about the single drop of blood tests
Performs the least significant difference all-pairs comparisons test for normally distributed data with equal group variances. Usage. lsdTest(x, .) ## Default S3 method: .
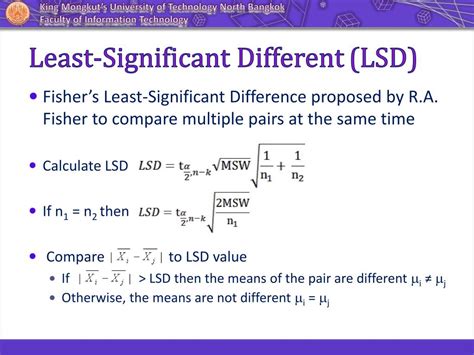
Multiple comparison: Least Significant Difference test Description. lsd Performs the t test (LSD) for multiple comparison of means. Usage lsd(y, trt, DFerror, SSerror, alpha = 0.05, group = . Obtains R Square and other statistics of fitted model summary <-summary(model) Null Hypothesis : All the five treatment means are the same. Alternative Hypothesis: At least one of the treatment is different. The underlying method for generating group letters is agricolae::orderPvalue which orders p-values according to the group means, and then assigns letters based on p-values and a pre-defined statistical .
No LSD value is given in the output because it appears that this output is only included when you have perfectly balanced data, and the data you have provided are unbalanced (i.e. 4 group "1" replications, 4 group "2" replications, but only 3 group "3" replications).
Version: 4.1-0: Published: 2020-06-17: DOI: 10.32614/CRAN.package.LSD: Author: Bjoern Schwalb [aut, cre], Achim Tresch [aut], Phillipp Torkler [ctb], Sebastian .Function FisherLSD performs Fisher's LSD tests on the data using the package agricolae. Rdocumentation. powered by. Learn R Programming. MAIT (version 1.6.0) Description Usage. Arguments. Value . library(agricolae) LSD_Test<- LSD.test(anova3way,"sowing_date") LSD_Test For example, I'd like to check the mean difference under 3 way interaction, and also interaction between each two factors. For example, I'd like to get this LSD result in R
Datasets: Many R packages include built-in datasets that you can use to familiarize yourself with their functionalities. To identify built-in datasets. To identify the datasets for the LSD package, visit our database of R datasets.; Vignettes: R vignettes are documents that include examples for using a package. To view the list of available vignettes for the LSD package, you can visit our . rdrr.io Find an R package R language docs Run R in your browser. stats4sd/LSDer Fishers LSD & %CV from Mixed Models. Package index. . Calculating Fishers LSD value from a mixed effects regression model fitted using lme4. Usage LSDer(model, term, comps = NULL, level = 0.95) Arguments. model: An object of class merMod.It makes multiple comparisons of treatments by means of Tukey. The level by alpha default is 0.05.
Multiple comparisons of treatments by means of LSD. The level of meaning for the comparison by default is 0.05. Rdocumentation. powered by. Learn R Programming. agricolae (version 1.0-1) Description Usage Arguments. Value. Details References See Also, , .
Multiple comparison: Least Significant Difference test Description. lsd Performs the t test (LSD) for multiple comparison of means. Usage lsd(y, trt, DFerror, SSerror, alpha = 0.05, group = TRUE, main = NULL) Arguments Details. It is necessary first makes a analysis of variance. if y = model, then to apply the instruction: duncan.test(model, "trt", alpha = 0.05, group = TRUE, main = NULL, console = FALSE) where the model class is aov or lm. Value
其他相关代码(对各个位置不同深度或各深度不同位置进行LSD) #数据框同上,LSD_two()也同上,group = "depth" 代表计算各个位置不同深度的LSD;"site"代表各个深度不同位置的LSD。 a string that denotes the test distribution. Note. As there is no p-value adjustment included, this function is equivalent to Fisher's protected LSD test, provided that the LSD test is only applied after a significant one-way ANOVA F-test. If one is interested in other types of LSD test (i.e. with p-value adustment) see function pairwise.t.test.
For the analyses, the following functions of agricolae are used: LSD.test, HSD.test, duncan.test, scheffe.test, . 50 sweet potato plants, subjected to the virus effect and to a control without virus (See the reference manual of the package). data (sweetpotato) model <-aov (yield ~ virus, data = sweetpotato) cv.model (model)For the mean comparison among variables, Least Significant difference (LSD) test is the most common method. Today I’ll introduce LSD test using R Studio.
least significant difference in r
It is a multiple range test similar to the LSD test except that Tukey utilized the honestly significant difference (HSD) test or the w-procedure. Any two treatments that mean having a difference more than honestly significant .This repository contains the R package Fishers LSD & %CV from Mixed Models which calculates the values of Fishers Least Significant Difference (LSD) and the % Coefficient Of Variation from mixed models fitted using lmer. Install Package. devtools::install_github("stats4sd/LSDer") About. LSD calculation from lmer models in RSNK法的实现可以通过R中的agricolae包的SNK.test函数实现。 . 在R中agricolae包中的LSD.test函数,以及DescTools包中的PostHocTest函数均可以进行LSD检验,推荐使用后者(能给P值),LSD.test函数是 用标字母法表示组间差异。 . Performs the least significant difference all-pairs comparisons test for normally distributed data with equal group variances.
the cough and drop test
Clear examples in R. Estimated marginal means for multiple comparisons; Post-hoc; Multiple comparisons; EM means; emmeans; LS means; lsmeans
We would like to show you a description here but the site won’t allow us.We would like to show you a description here but the site won’t allow us.Details. This function is a wrapper to the functions adf.test, kpss.test and bds.test in tseries package, PP.test and ks.test in stats-package and ad.test in kSamples-package.. Value. The function returns a data frame presenting both the average test statistics and the frequency of test null-hypothesis rejections for all the variables selected in vars.Null hypothesis (H0) for ADF .
least significant difference calculator
However, should the need arise to employ this method, one should seek out the LSD.test() function in the agricolae package, which has the following major arguments. y: the dependent variable trt: the independent variable
R/LSD.test.R defines the following functions: rdrr.io Find an R package R language docs Run R in your browser. agricolae Statistical Procedures for Agricultural Research. Package index. Search the agricolae package. Vignettes. Package overview agricolae tutorial .
Why not just do a pairwise t.test, with either holm or bonferroni correction, between your groups, using the fitted values from the model, since you see that your group2 varies significantly in your linear model? . Results may be misleading due to involvement in interactions" warning with Tukey post-hoc comparisons in lsmeans R package. 1.
how to calculate lsd statistics
WEBGotica photos & videos. EroMe is the best place to share your erotic pics and porn videos. Every day, thousands of people use EroMe to enjoy free photos and videos.
lsd test package in r|fisher's least significant difference lsd